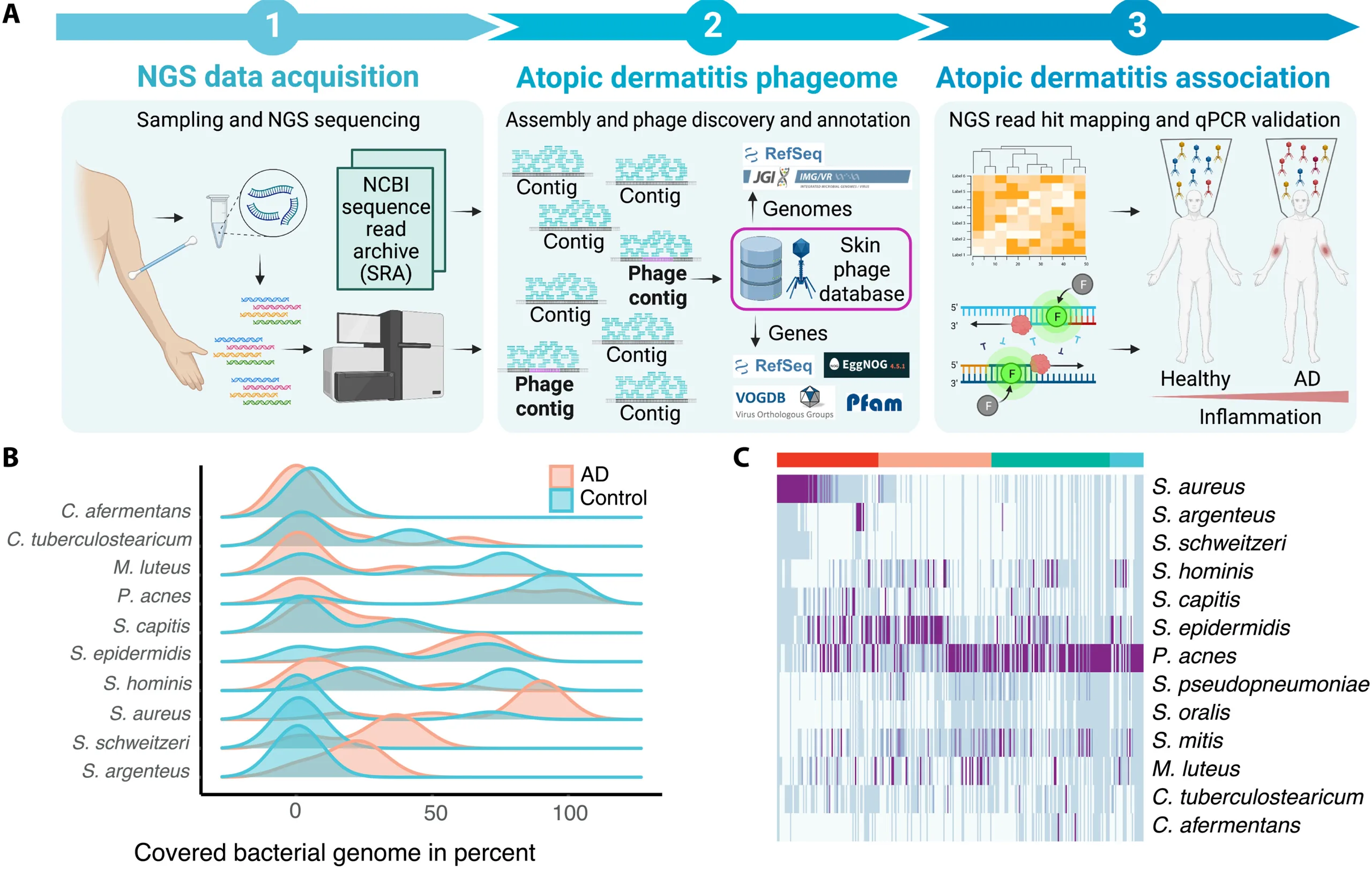
Study workflow and bacterial communities in healthy skin and AD.
In this study published in Science Advances, Wolfgang Weninger and his team from the Medical University of Vienna, have highlighted the role of bacteriophages in the skin’s microbiome, particularly in relation to atopic dermatitis (AD).
By analyzing skin swabs from both healthy individuals and AD patients using shotgun metagenomics, the researchers identified over 13,000 potential viral DNA sequences. These sequences were combined into 164 putative viral genomes, 133 of which were identified as bacteriophages.
Interestingly, the study found that the diversity of these viral genomes, measured by the Shannon diversity index, did not correlate with the presence of AD. This suggests that the sheer variety of viruses isn’t the main factor in the disease’s progression. However, the research did uncover 28 viral genomes that significantly differed between healthy and AD-affected skin, indicating distinct viral communities in inflamed skin.
Moreover, the presence of these viral genomes was shown to be independent of the abundance of their bacterial hosts. This means that changes in the phageome aren’t just a byproduct of bacterial changes but may play a direct role in skin health and disease.
These findings suggest that shifts in the skin’s viral community could drive changes in bacterial populations, contributing to the development of AD. This opens up new possibilities for therapies that target both the viral and bacterial components of the skin microbiome.
Wolfgang Weninger will be discussing these insightful findings at the 7th World Conference Targeting Phage Therapy, taking place on June 20-21 in Malta. Learn more.
Photo Credits: M. Wielscher et al. Sci. Adv. (2023)
