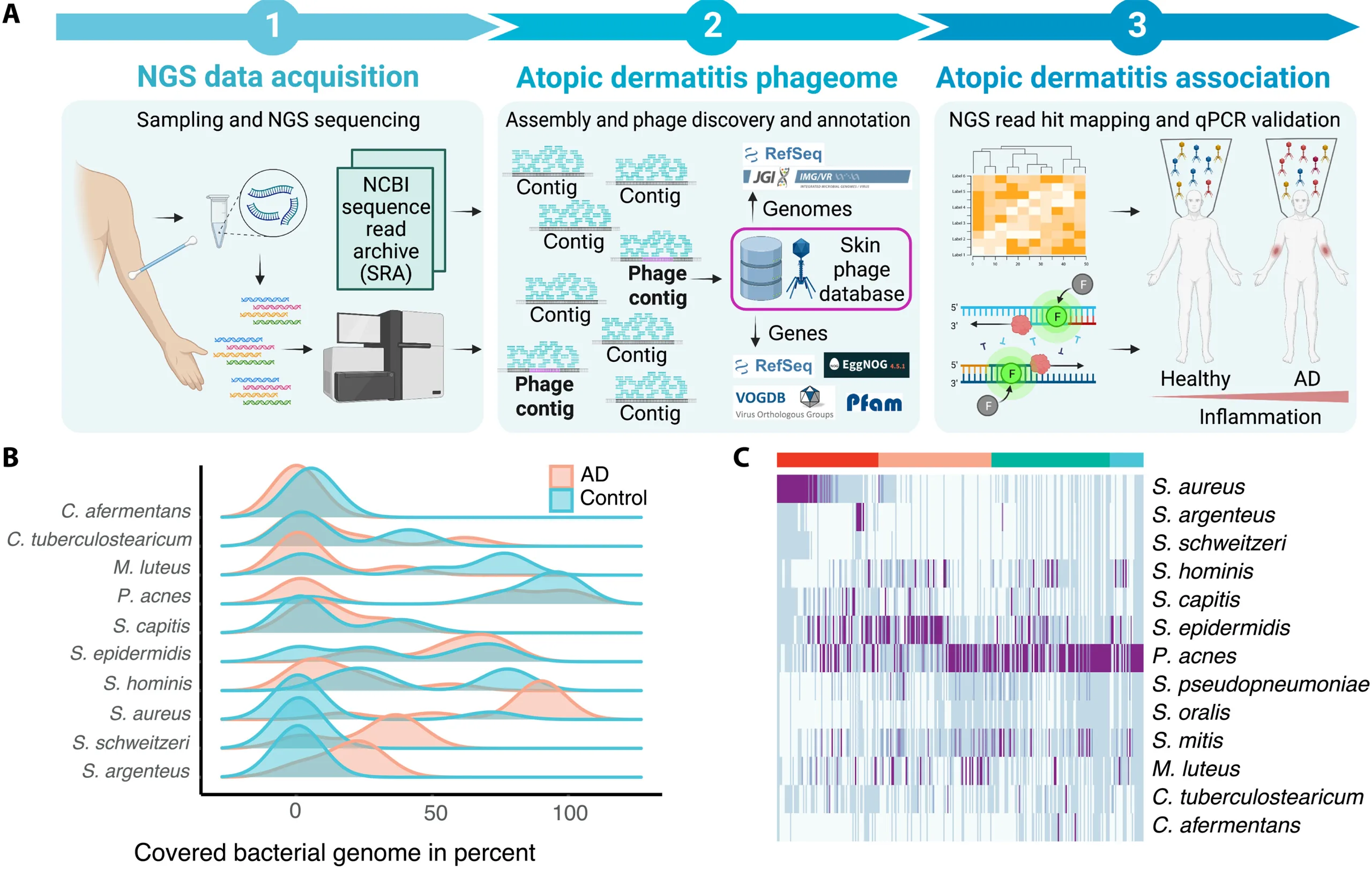
Scientists have uncovered new details about viruses that live inside bacteria in the human gut, challenging long held assumptions about how dynamic the microbiome really is.
Using advanced long read DNA sequencing, researchers tracked viral DNA embedded within gut bacteria in healthy adults over a two year period. These viruses, known as prophages, are bacteriophages that insert their genetic material into bacterial genomes.
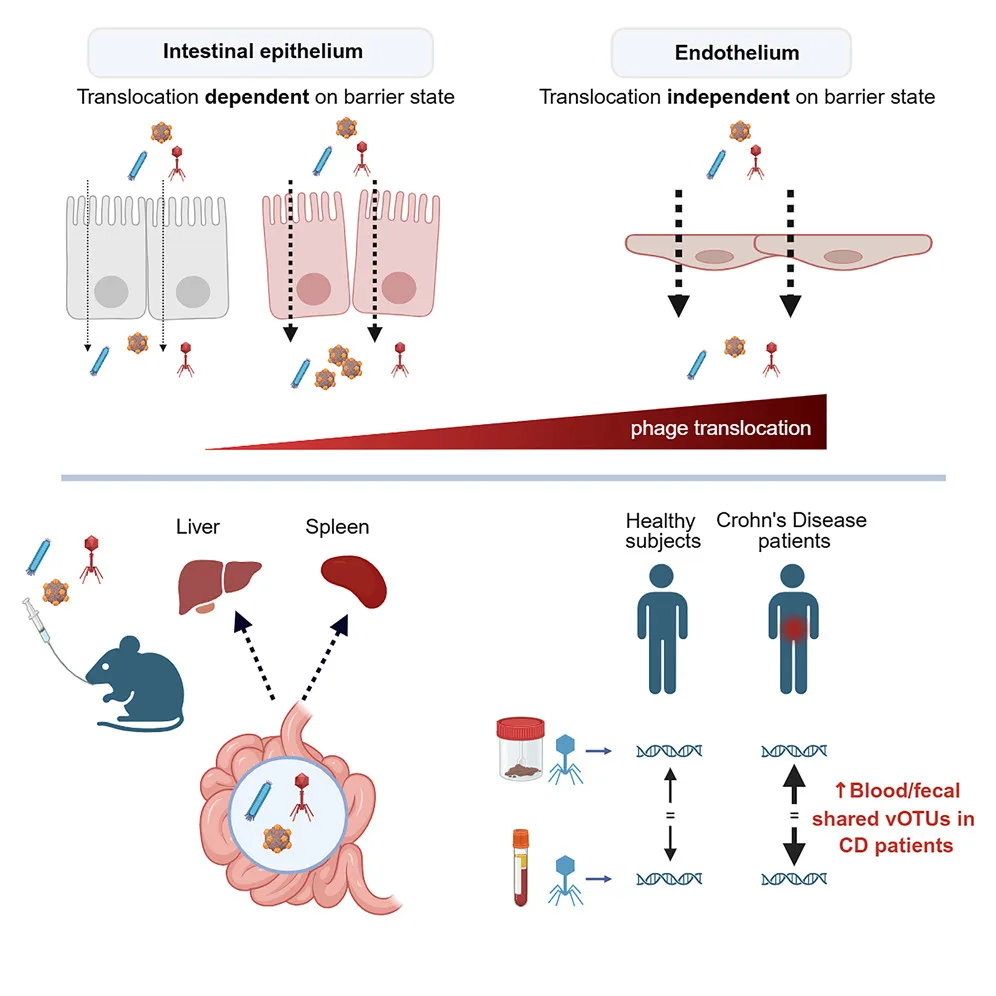
The study found that most gut viruses are remarkably stable, remaining inside the same bacterial hosts for years. Only a small fraction were gained or lost over time, suggesting that the gut microbiome is more steady and predictable than previously thought.
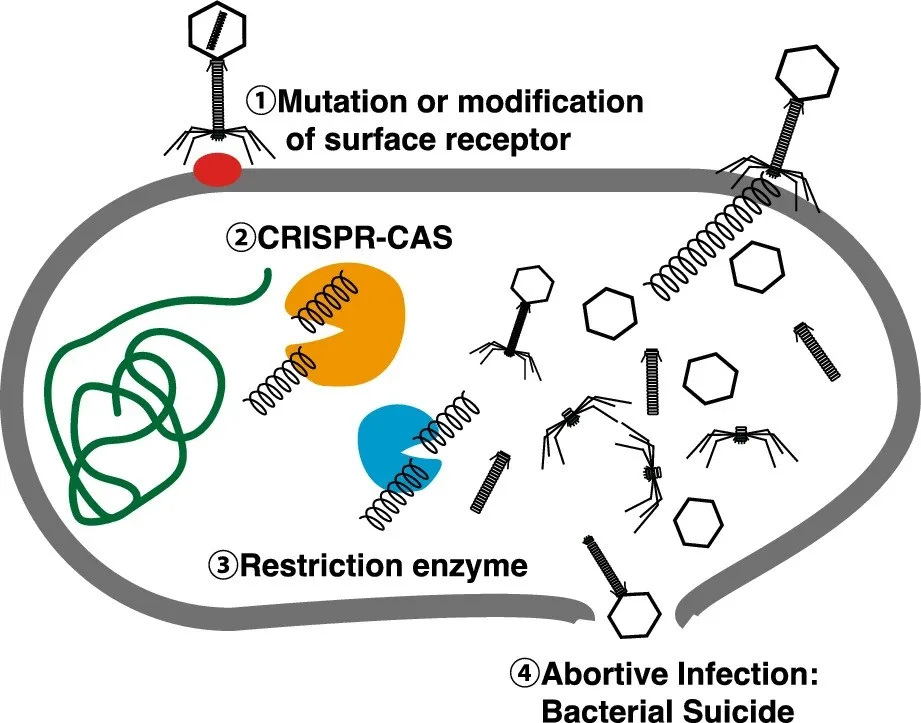
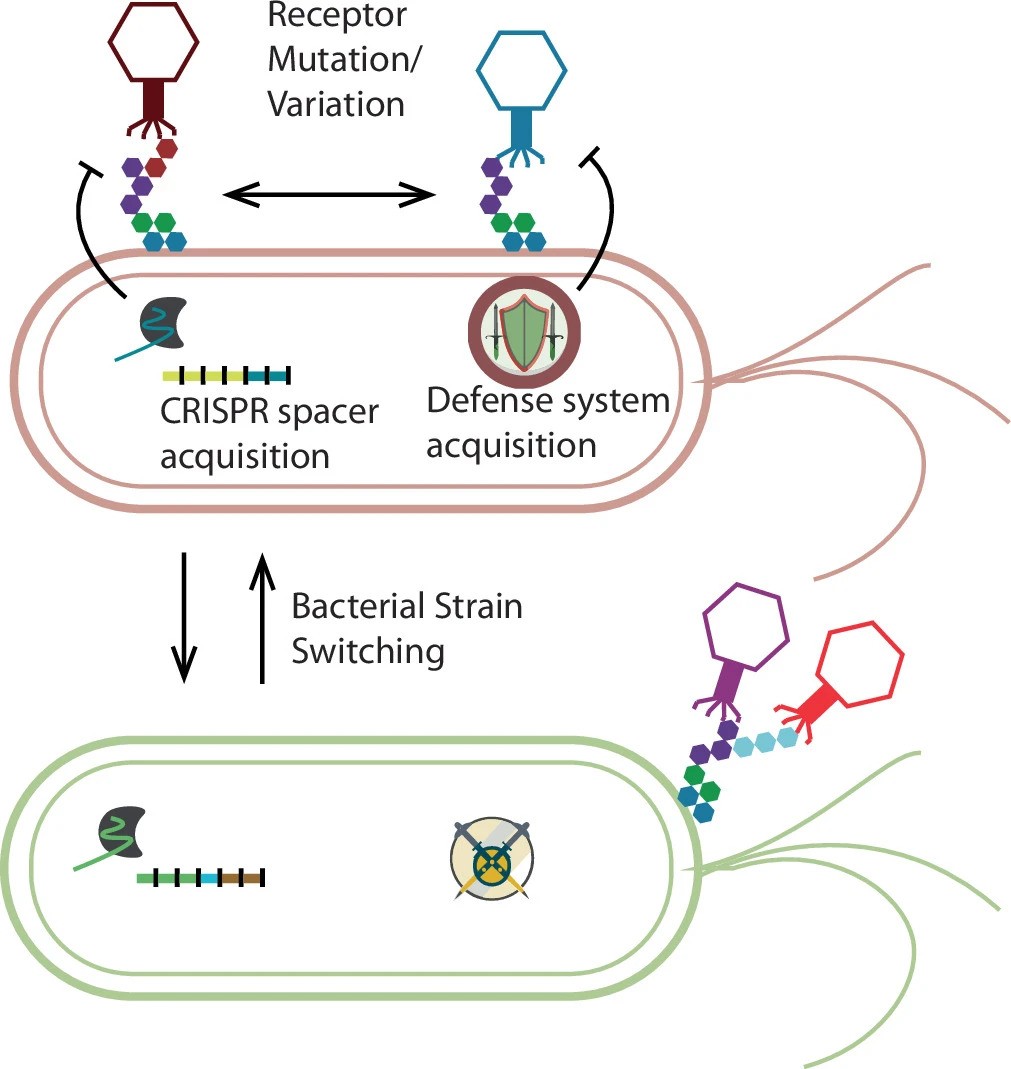
Researchers also discovered that some viruses can live in multiple types of bacteria, contradicting the idea that phages are usually limited to a single host.
A key finding was the discovery of a new group of prophages called IScream phages. These viruses use bacterial mobile DNA elements to insert and remove themselves from host genomes, revealing a previously unknown mechanism of viral integration. This suggests gut viruses may be more genetically flexible than scientists realized.
The findings were made possible by long read sequencing technology, which allows scientists to assemble complete bacterial genomes and precisely locate viral DNA, something earlier methods could not do reliably.
Experts say the work reshapes understanding of how viruses influence gut health and could have long term implications for microbiome based therapies, including treatments targeting digestive, immune, and metabolic diseases.
Click here to read more
We are pleased to announce that the 9th Annual Meeting Targeting Phage Therapy 2026 will be held in Valencia, Spain, on June 11–12. We look forward to welcoming you.