Bacteriophage T4 has enormous potential for biomedical applications due to its large size, capsid architecture, and high payload capability for protein and DNA delivery.
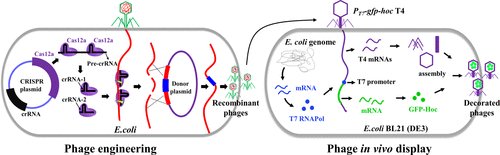
However, it is not very easy to genetically engineer its genome heavily modified by cytosine hydroxymethylation and glucosylation. The glucosyl hydroxymethyl cytosine (ghmC) genome of phage is completely resistant to most restriction endonucleases and exhibits various degrees of resistance to CRISPR-Cas systems.
In this study, Dong et al. found that the type V CRISPR-Cas12a system, which shows efficient cleavage of ghmC-modified genome when compared to the type II CRISPR-Cas9 system, can be synergistically employed to generate recombinant T4 phages.
Focused on surface display, they analyzed the ability of phage T4 outer capsid proteins Hoc (highly antigenic outer capsid protein) and Soc (small outer capsid protein) to tether, in vivo, foreign peptides and proteins to T4 capsid. The obtained data showed that while these could be successfully expressed and displayed during the phage infection, shorter peptides are present at a much higher copy number than full-length proteins. However, the copy number of the latter could be elevated by driving the expression of the transgene using the strong T7 RNA polymerase expression system.
This CRISPR-inspired approach has the potential to expand the application of phages to various basic and translational research projects.
Read more about this phage engineering approach.
Targeting Phage Therapy 2023 will introduce you to the latest discoveries in phage engineering. Secure a place.
Targeting Phage Therapy 2023
6th World Conference
June 1-2, 2023 – Paris, France
