Processing workflow. Credits: Flinders Accelerator for Microbiome Exploration, College of Science and Engineering, Flinders University
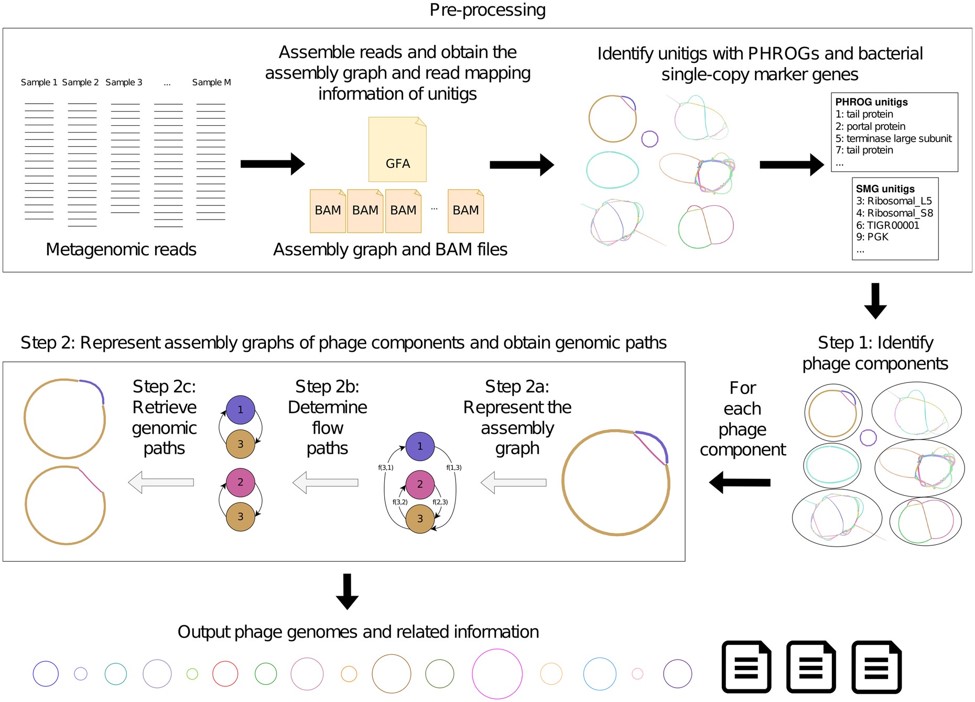
In a significant leap forward for phage research, Flinders University’s College of Science and Engineering introduces “Phables”, a bioinformatics software program designed to address the challenges in identifying and characterizing phage genomes within fragmented viral metagenome assemblies. This innovative tool, inspired by the abstract’s motivation and results, marks a significant advancement in phage research.
Microbial communities profoundly influence human health and various environments, with bacteriophages or phages playing a crucial role in modulating bacterial communities. High-quality phage genome sequences are vital for advancing our understanding of phage biology, conducting comparative genomics studies, and developing phage-based diagnostic tools. However, existing viral identification tools often face challenges in viral assembly, resulting in genome fragmentation and incomplete recovery of phage genomes.
Phables tackles this issue by employing a novel computational method. It identifies phage-like components in the assembly graph, models each component as a flow network, and utilizes graph algorithms and flow decomposition techniques to identify genomic paths. Experimental results from viral metagenomic samples collected from different environments demonstrate that Phables outperforms existing tools, recovering over 49% more high-quality phage genomes on average.
Importantly, Phables has the capability to resolve variant phage genomes with over 99% average nucleotide identity—a feat that existing tools struggle to achieve. This ability to make fine distinctions contributes to a more comprehensive understanding of phage diversity and evolution.
Learn more about this fascinating tool.
