A paper by Robert A Edwards and Bhavya Papudeshi from Flinders University, focused on refining the understanding and application of bacteriophages. Bacteriophages are viruses that infect bacteria, exerting a significant influence on microbial communities and ecosystems. They have garnered considerable attention for their potential role in combating antibiotic resistance.
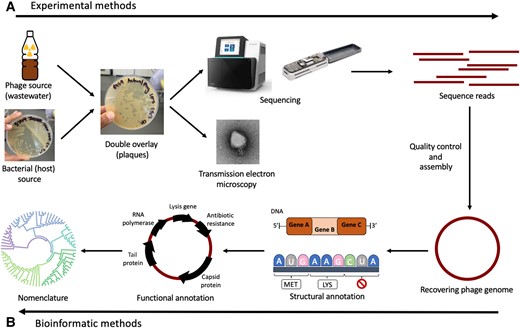
Phage therapy predominantly relies on lytic phages due to their efficacy in killing bacteria, while temperate phages, capable of transferring antibiotic resistance or toxin genes, are typically avoided. The selection process hinges on assessing plaque morphology and conducting genome sequencing to ensure the safety and effectiveness of the chosen phages.
The study underscored the importance of accurately annotating phage genomes, identifying crucial genomic features, and assigning functional labels to protein-coding sequences. These annotations play a pivotal role in preventing the inadvertent transfer of undesirable genes, such as those associated with antimicrobial resistance or toxins, during phage therapy procedures.
Furthermore, the review delved into the International Committee on Taxonomy of Viruses (ICTV), which provides a standardized phage nomenclature system aimed at simplifying classification and communication within the scientific community. By adhering to this established framework, researchers can enhance the accuracy and clarity of their findings, facilitating more effective collaboration and knowledge dissemination.
Accurate phage genome annotation and nomenclature not only offer valuable insights into phage-host interactions, replication strategies, and evolutionary patterns but also expedite advancements in understanding phage diversity and evolution. Moreover, they play a pivotal role in driving the development of innovative phage-based therapies, offering promising solutions to the challenges posed by antibiotic resistance.
Image Description:
