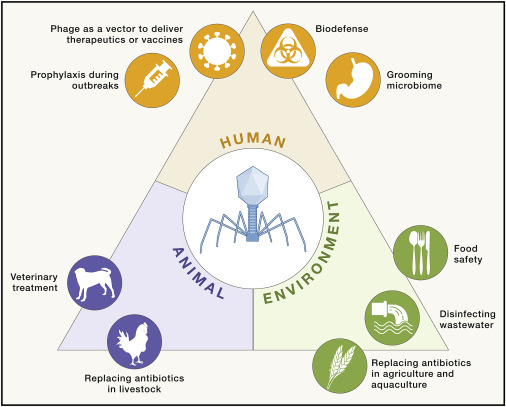
Researchers from the Max Planck Institute for Evolutionary Biology have introduced an innovative approach to strengthen the effectiveness of phage therapy against drug-resistant bacteria. As antibiotic resistance continues to rise globally, finding alternative treatments is increasingly urgent, and phages – viruses that target bacteria – have emerged as a promising option.
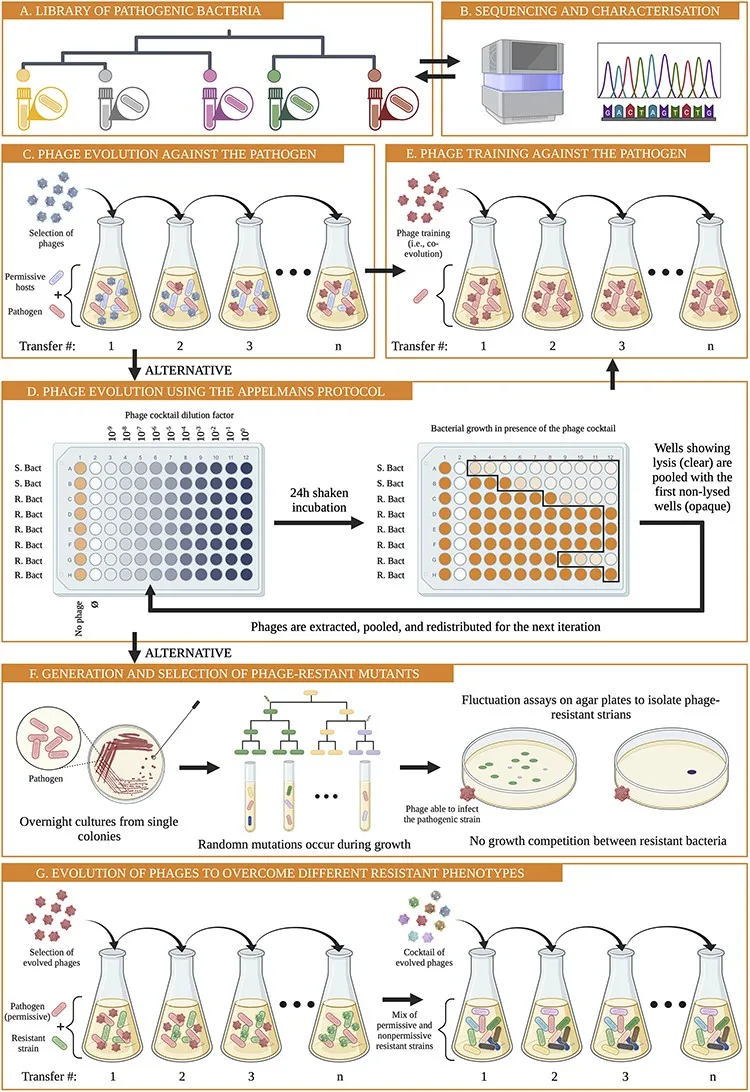
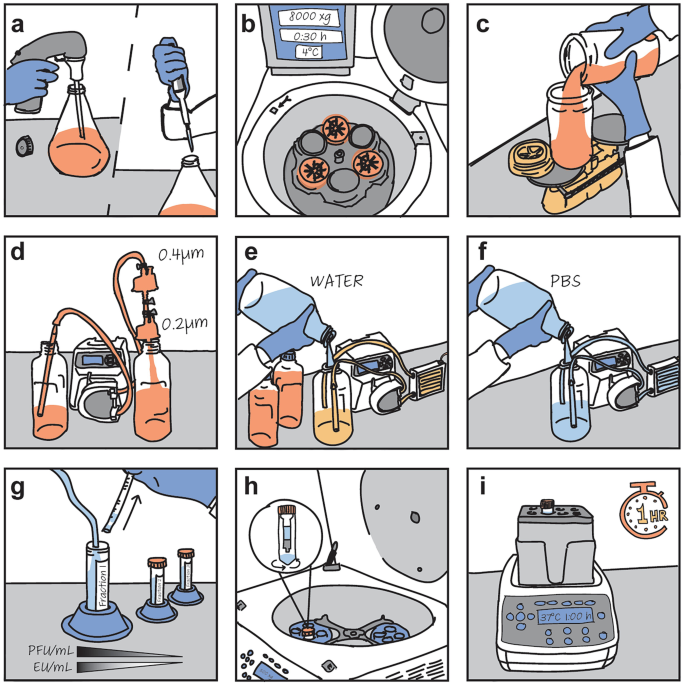
The proposed procedure to develop a phage model system into a therapeutic agent.
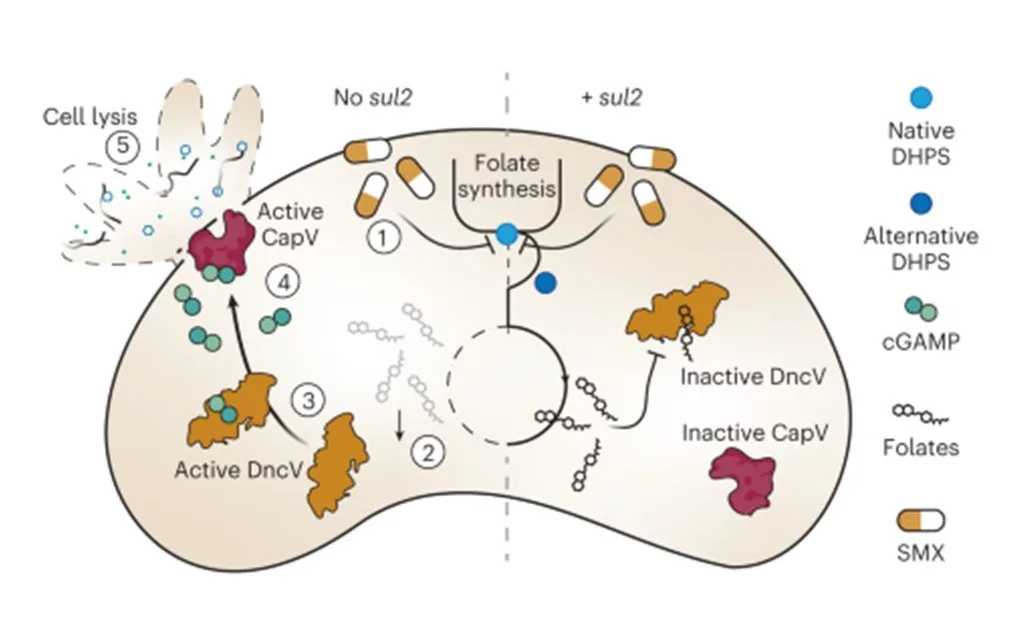
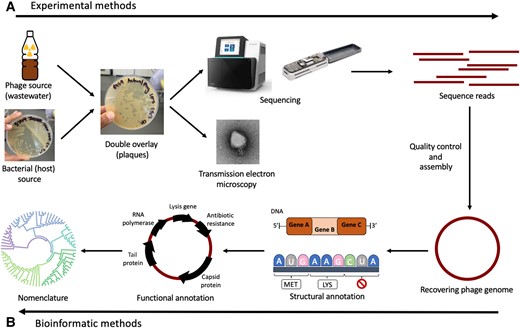
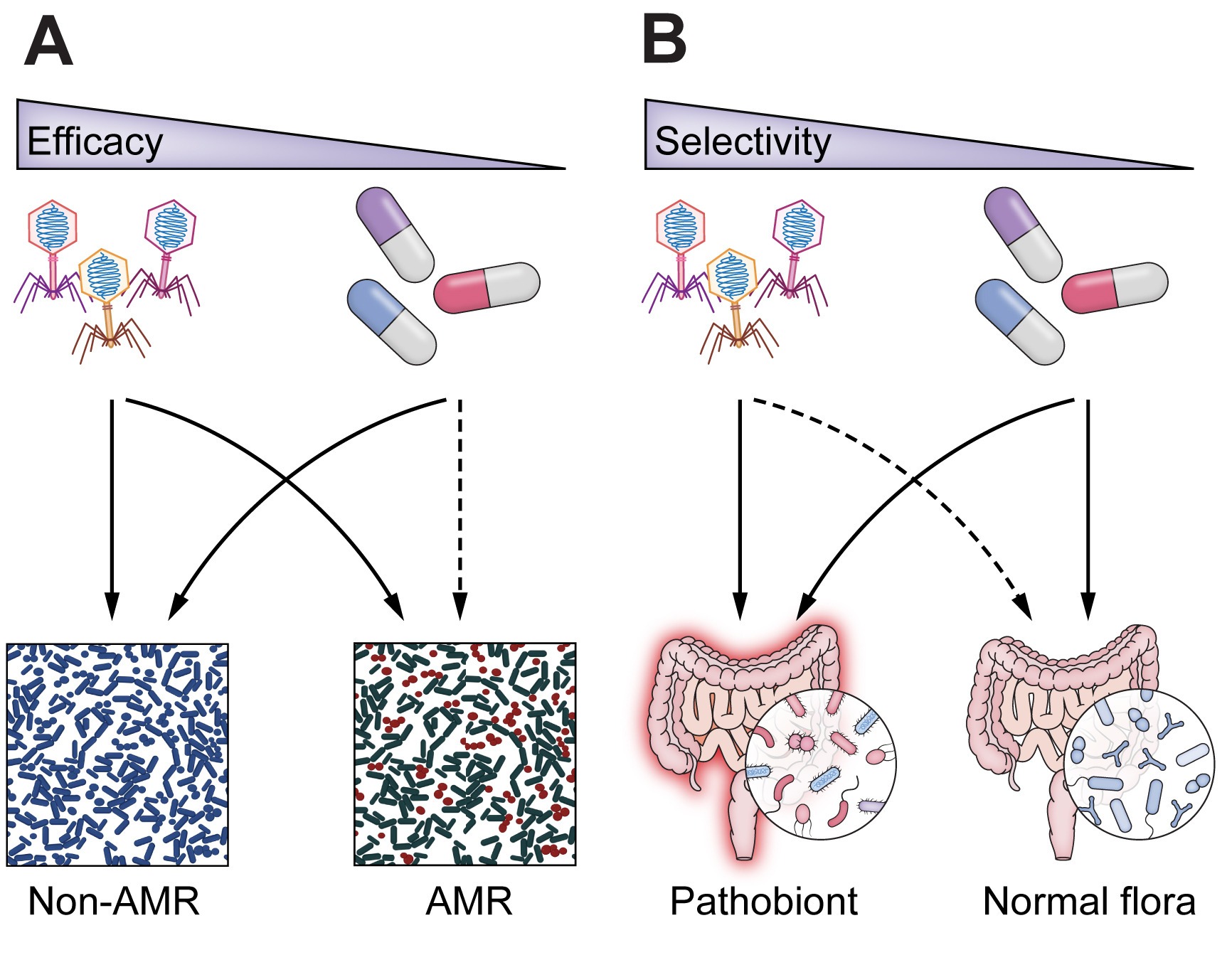
The study emphasizes the importance of well-established phage model systems, which have been extensively studied over the years, for safe and efficient treatment. However, these models have faced challenges in directly combating harmful bacteria.
Nevertheless, the researchers offer a solution. By modifying these model phages through genetic engineering or evolution, they can be adapted to target harmful bacteria more effectively. This breakthrough could pave the way for personalized phage treatments tailored to specific infections.
These findings represent a significant advancement in the fight against drug-resistant infections.
Dr. Frederic Bertels, lead author of this study, will join us Malta for Phage Therapy 2024 this June to further elaborate these findings.
Photo credits: Romeyer Dherbey, J., & Bertels, F. (2024). Virus Evolution, 10(1), veae007.