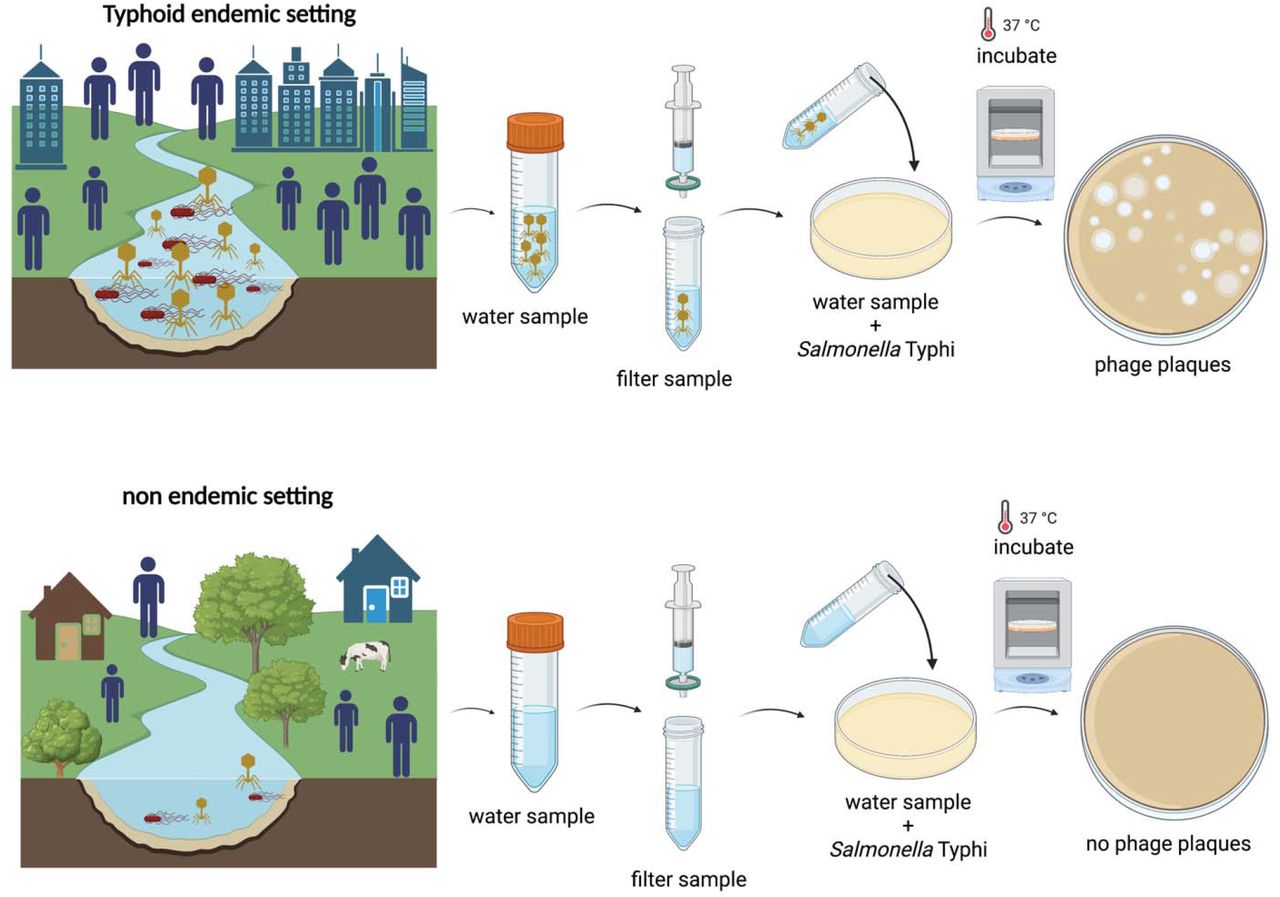
Environmental surveillance, using detection of Salmonella Typhi DNA, has emerged as a potentially useful tool to identify typhoid-endemic settings; however, it is relatively costly and requires molecular diagnostic capacity. Shrestha et al. sought to determine whether S. Typhi bacteriophages are abundant in water sources in a typhoid-endemic setting, using low-cost assays.
They collected drinking and surface water samples from urban, peri-urban and rural areas in 4 regions of Nepal. They performed a double agar overlay with S. Typhi to assess the presence of bacteriophages. They isolated and tested phages against multiple strains to assess their host range. They performed whole genome sequencing of isolated phages, and generated phylogenies using conserved genes.
S. Typhi-specific bacteriophages were detected in 54.9% of river water samples and 6.3% drinking water samples from the Kathmandu Valley and Kavrepalanchok. Water samples collected within or downstream of population-dense areas were more likely to be positive (72.6%, 193/266) than those collected upstream from population centers (5.3%, 5/95). In urban Biratnagar and rural Dolakha, where typhoid incidence is low, only 6.7% (1/15, Biratnagar) and 0% (0/16, Dolakha) samples contained phages.
All S. Typhi phages were unable to infect other Salmonella and non-Salmonella strains, nor a Vi-knockout S. Typhi strain. Representative strains from S. Typhi lineages were variably susceptible to the isolated phages. Phylogenetic analysis showed that S. Typhi phages belonged to two different viral families (Autographiviridae and Siphoviridae) and clustered in three distinct groups.
S. Typhi bacteriophages were highly abundant in surface waters of typhoid-endemic communities but rarely detected in low typhoid burden communities. Bacteriophages recovered were specific for S. Typhi and required Vi polysaccharide for infection.
Screening small volumes of water with simple, low-cost plaque assays enables detection of S. Typhi phages and should be further evaluated as a scalable tool for typhoid environmental surveillance.
In summary:
-
Typhoid phages are detectable in surface water using simple assays, in communities with high typhoid burden.
-
Bacteriophages are highly specific for S. Typhi and required Vi polysaccharide for infection.
-
S. Typhi phages have a broad lytic activity against the S. Typhi strains circulating in Nepal.
-
Phage plaque assay can be used as a low-cost tool to identify communities where typhoid is endemic.
-
The high abundance of phages in river water suggest that this could be an alternative to molecular methods for environmental surveillance for typhoid.
Targeting Phage Therapy 2023 will introduce you to the latest discovered phages and phage related breakthroughs. Learn about the program.
Targeting Phage Therapy 2023 Congress
6th World Conference
June 1-2, 2023 – Paris, France
