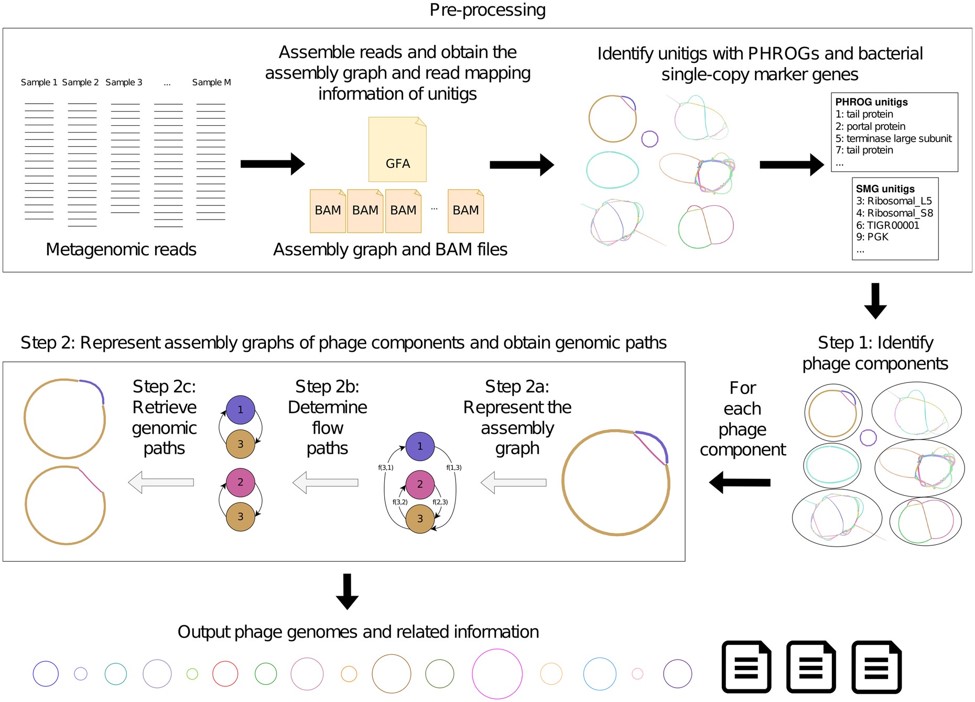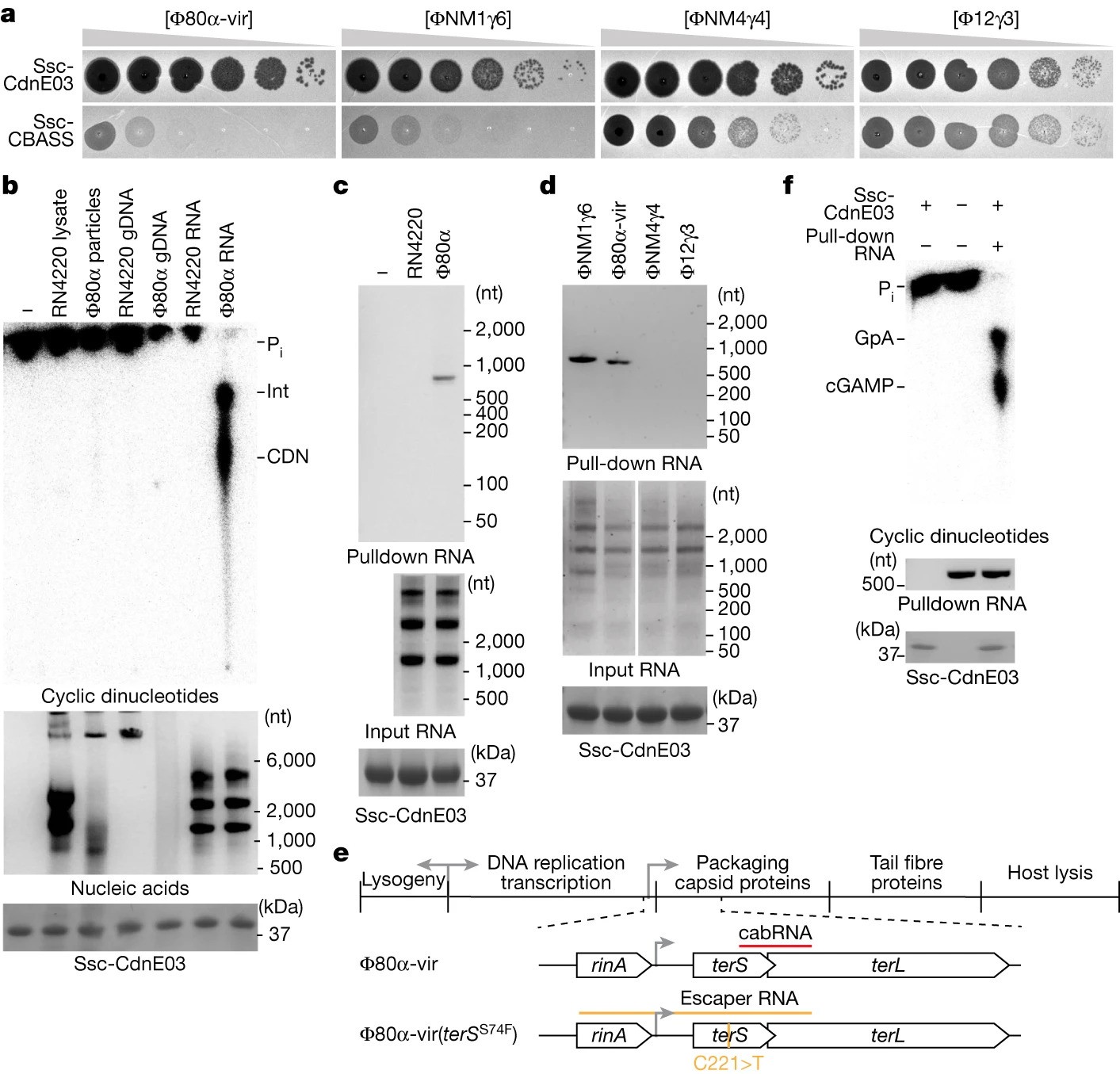
A viral RNA produced during Φ80α-vir infection activates Ssc-CdnE03 in vitro
Researchers at The Rockefeller University’s Laboratory of Bacteriology have uncovered a novel mechanism through which bacteria defend against viral infections, known as the cyclic oligonucleotide-based antiphage signaling system (CBASS). This intricate immune response protects prokaryotes by triggering the production of cyclic oligonucleotides, activating proteins that induce the death of infected host cells.
Key Findings:
- Staphylococcal phages produce a structured RNA called CBASS-activating bacteriophage RNA (cabRNA) from terminase subunit genes.
- CabRNA binds to the CdnE03 cyclase, promoting cGAMP synthesis and activating the CBASS immune response to prevent phage infection.
- Phages evading CBASS defense harbor mutations leading to longer cabRNA, hindering activation and revealing insights for combating antibiotic resistance.
- Parallels exist between CBASS and animal immune responses, highlighting shared viral sensing mechanisms across diverse life domains.
- CBASS cyclases, central to bacterial immune responses, are considered ancient relatives of animal cyclic GMP-AMP synthase (cGAS).
- Unlike animal cGAS sensing viral DNA, CBASS in bacteria detects specific RNA structures, identified as cabRNA during phage infection.
Future Research:
- Ongoing investigations will delve into cabRNA’s characteristics and its role in phage infection, potentially yielding breakthroughs in understanding bacterial defense systems.
Understanding bacterial defense mechanisms, especially CBASS, holds promise in addressing antibiotic resistance. The discovery suggests using specific phages that do not trigger CBASS responses, offering a potential avenue for future research in combating antimicrobial-resistant bacteria and advancing bacterial infection treatments.